Mapping protein sequences to their biological functions is crucial in biology, as proteins perform diverse roles in organisms. Functions are categorized using ontologies like Gene Ontology (GO) terms, Enzyme Commission (EC) numbers, and Pfam families. Computational predictions are essential due to the cost of lab experiments and rapid database growth. Techniques include homology-based methods, which use sequence alignment tools like BLAST to infer function, and deep learning methods, which predict functions directly from sequences. Challenges include generalizing predictions to new protein classes and dealing with proteins that lack similarity to known sequences, known as the “dark matter” of the protein universe.
Researchers from Google DeepMind, Google, and the University of Cambridge introduced ProtEx, a retrieval-augmented method for protein function prediction. ProtEx uses exemplars from a database to enhance accuracy, robustness, and generalization to new classes. It combines non-parametric similarity searches with deep learning inspired by retrieval-augmented techniques in NLP and vision. ProtEx retrieves positive and negative exemplars using tools like BLAST and trains a neural model to compare these exemplars with the query. This approach achieves state-of-the-art results in predicting EC numbers, GO terms, and Pfam families, particularly excelling with rare and dissimilar sequences. Ablation studies confirm the efficacy of the pretraining strategy and exemplar conditioning.
ProtEx builds on traditional protein similarity searches and recent neural models for protein function prediction. Conventional methods, like BLAST, retrieve homologous sequences to infer functions. Deep learning models, however, can outperform these by mapping sequences directly to functions. ProtEx integrates these approaches, using BLAST to retrieve exemplars and a neural model to condition predictions on these exemplars. This method excels, especially for rare and unseen classes. Retrieval-augmented models inspire it in NLP and vision, which enhance performance by incorporating context from retrieved exemplars. ProtEx effectively adapts to new labels without additional fine-tuning, leveraging multi-sequence pretraining for improved prediction accuracy.
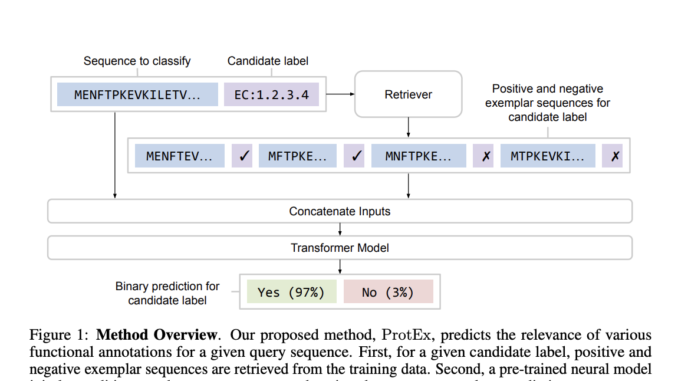
ProtEx aims to predict protein function labels for a given amino acid sequence. The process involves retrieving relevant positive and negative exemplar sequences for each candidate label using methods like BLAST. The model predicts the relevance of each label by conditioning on the sequence and its exemplars and aggregates these predictions to form the final label set. A candidate label generator reduces the number of labels considered to improve efficiency. Pre-training involves comparing sequence pairs with varying similarities while fine-tuning uses training data to create positive and negative examples. The model employs a T5 Transformer architecture to handle these tasks.
ProtEx was evaluated using several datasets on EC number, GO term, and Pfam classification tasks. BLAST was used as the retriever for EC and GO tasks, while a per-class retrieval approach was applied to the larger Pfam dataset. In EC and GO prediction tasks, ProtEx outperformed previous methods and showed significant improvements when conditioned on exemplar sequences. ProtEx also achieved state-of-the-art performance on the Pfam dataset, demonstrating consistent accuracy across common and rare protein families. The model was pre-trained on sequence pairs and fine-tuned with both positive and negative exemplars using a T5 Transformer architecture.
In conclusion, ProtEx introduces a method that integrates homology-based similarity search with pre-trained neural models, achieving state-of-the-art results in EC, GO, and Pfam classification tasks. Despite the increased computational requirements due to encoding multiple sequences and making independent class predictions, efficiency improvements are possible through architectural adjustments and candidate label generation. Future enhancements could leverage advanced similarity search techniques and specialized architectures. While the method enhances protein function predictions, verification through wet lab experiments remains essential for critical applications. This approach builds on existing tools, offering more accurate and robust functional annotations of proteins.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 43k+ ML SubReddit | Also, check out our AI Events Platform
![]()
Sana Hassan, a consulting intern at Marktechpost and dual-degree student at IIT Madras, is passionate about applying technology and AI to address real-world challenges. With a keen interest in solving practical problems, he brings a fresh perspective to the intersection of AI and real-life solutions.




Be the first to comment